
- This module contains a series of novel parallel algorithms to perform suffix-sorting and BWT construction of very large texts and text collections.
- For example, the single string BWT construction can be used to index the whole human genome in under 2 minutes on a Tesla K20 GPU with 5GB of device memory and 16GB of system memory - while the string-set BWT construction algorithm has been tested with up to 500M x 100bp reads on a system with the same GPU and as little as 32GB of system memory.
- The functions are split into two groups: functions that operate on host-side strings and string-sets, and functions that operate on device-side strings and string-sets. The latter are grouped into the cuda namespace.
- The large string BWT construction uses a GPU implementation of J.Kaerkkaeinen's Blockwise Suffix Sorting framework, customized around a new GPU-based block sorter that employs a mixture of a novel, high-performance MSB radix sorting algorithm and a high-period DCS sorter. The resulting algorithm can sort strings containing several billion characters at up to 70M suffixes/s on a Tesla K40, and is practically insensitive to LCP length.
- The large string-set BWT construction algorithm is a new derivation, originally inspired by:
"GPU-Accelerated BWT Construction for Large Collection of Short Reads"
C.M. Liu, R.Luo, T-W. Lam
http://arxiv.org/abs/1401.7457
Performance
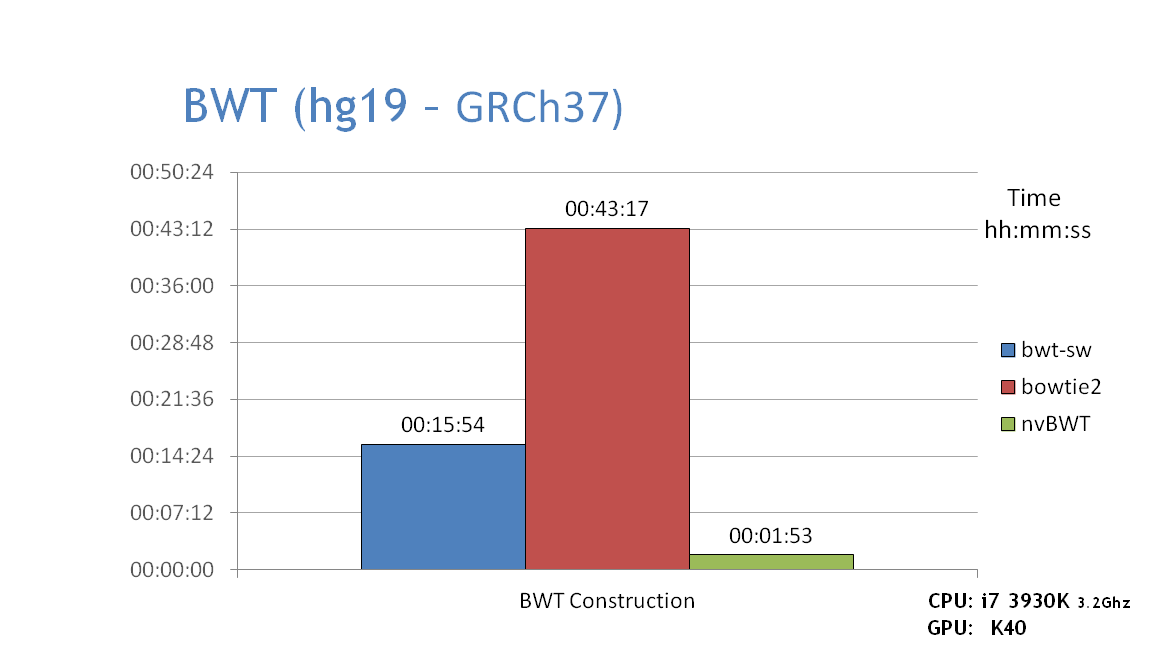
- The graph below shows NVBIO's BWT construction performance on the whole human genome compared to two other popular CPU based BWT builders:
Technical Overview
- A complete list of the classes and functions in this module is given in the Sufsort Module documentation.

 1.8.4
1.8.4