HEALPix Tutorial#
This tutorial demonstrates key HealPIX functionality including:
Visualization using double pixelization
Zonal averaging
Regridding between different grids
Reordering between different pixel ordering schemes
Padding for machine learning applications
Installation#
Earth2grid has some compiled components (in CUDA and C). So some care is required to install. First, install pytorch, and setuptools
# python3 -m venv .venv # make venv
# source .venv/bin/activate # activate venv
pip install torch setuptools
pip install --no-build-isolation https://github.com/NVlabs/earth2grid/archive/main.tar.gz
Warning
It is very important to use the --no-build-isolation flag. Otherwise, the library may be built against a different version of pytorch than you have installed, this will cause strange “missing symbols” errors.
Setup#
First, let’s import the necessary libraries and create a sample HealPIX grid.
#| label: setup
import matplotlib.pyplot as plt
import numpy as np
import torch
import earth2grid
from earth2grid import healpix
# Create a sample HealPIX grid
level = 3 # Resolution level (higher = finer resolution)
grid = healpix.Grid(level=level, pixel_order=healpix.PixelOrder.RING)
print(grid.lat[:5])
[84.14973294 84.14973294 84.14973294 84.14973294 78.28414761]
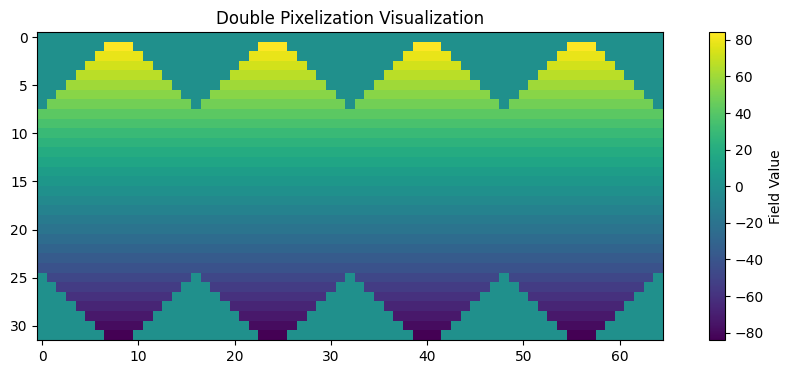
Visualization using Double Pixelization#
Double pixelization provides a visually appealing way to view HealPIX data without interpolation, preserving the native pixel structure. This is particularly useful for quick visualization with image viewers without distorting the native pixels of the image.
# Convert to double pixelization
field_double = healpix.to_double_pixelization(grid.lat)
plt.figure(figsize=(12, 4))
plt.imshow(field_double)
plt.colorbar(label='Field Value')
plt.title('Double Pixelization Visualization')
plt.show()
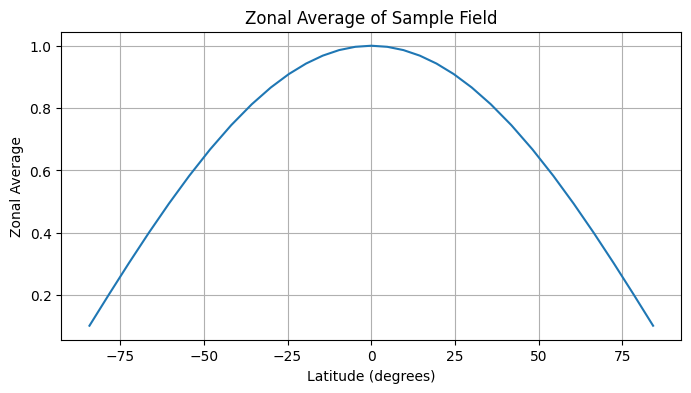
Zonal Averaging#
Zonal averaging computes the mean value of a field along latitude circles. This is useful for analyzing latitudinal patterns in the data.
# Compute the zonal average of the field
field = np.cos(np.deg2rad(grid.lat))
zonal_avg = healpix.zonal_average(field)
lat = healpix.zonal_average(grid.lat)
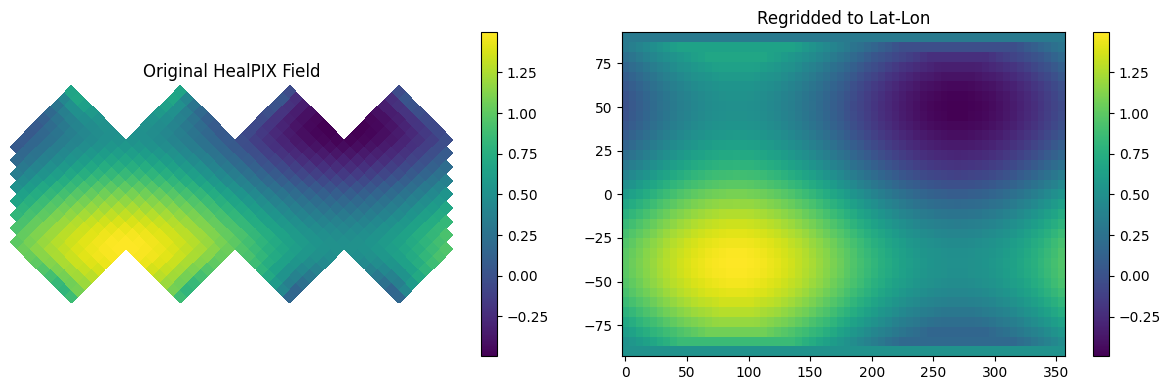
Regridding#
Regridding allows us to transform data between different grid types. Here we demonstrate two types of regridding:
From HEALPix to a regular lat-lon grid
Between HEALPix grids of different resolutions
Regridding to Lat-Lon#
earth2grid has builtin utilities for bilinear interpolation from any supported grid to arbitrary lat-lon points.
HEALPIX to Lat-Lon#
#| label: regrid-latlon
# Create a lat-lon grid
nlat, nlon = 33, 64
latlon_grid = earth2grid.latlon.equiangular_lat_lon_grid(nlat, nlon)
# Create regridder from HealPIX to lat-lon
regridder = earth2grid.get_regridder(grid, latlon_grid)
# Regrid the field
# must be a torch tensor
field = np.cos(np.deg2rad(grid.lat+ 40)) ** 2 + 0.5 * np.sin(np.deg2rad(grid.lon))
field_regridded = regridder(torch.from_numpy(field))
A more verbose way, but flexible, way to do this is using the .get_bilinear_regridder_to method.
#| label: regrid-latlon-verbose
lat = np.linspace(-90, 90, 33)
lon = np.linspace(0, 360, 64)
regridder = grid.get_bilinear_regridder_to(lat[:, np.newaxis], lon[np.newaxis, :])
field_regridded = regridder(torch.from_numpy(field))
print(field_regridded.shape)
torch.Size([33, 64])
Note
Exercise: use get_bilinear_regridder_to to regrid to a list of unstructured points.
The regridding objects (and most operations in earth2grid) are fully differentiable and GPU capable
x = torch.randn(grid.shape)
x.requires_grad_(True)
regridder.to(x.dtype) # need to make sure the regridder is the same type as the input
y = regridder(x)
(y ** 2).sum().backward()
print(x.grad[:5])
tensor([ 21.8016, -15.9544, -21.5755, 14.5128, 9.7041])
Lat-Lon to HEALPIX#
from earth2grid import latlon
llgrid = latlon.LatLonGrid(lat=lat, lon=lon)
hpxgrid = healpix.Grid(level=6)
regridder = llgrid.get_bilinear_regridder_to(hpxgrid.lat, hpxgrid.lon)
field = lat[:, None] + 0 * lon # trick to get (nlat, nlon) array
out = regridder(torch.from_numpy(field))
print(out.shape)
torch.Size([49152])
HEALPix Pixel Orders#
earth2grid supports three main pixel ordering schemes:
RING: Pixels are ordered in rings of constant latitude
NEST: Pixels are ordered in a hierarchical, nested pattern
XY: Pixels are ordered in a Cartesian-like grid within each face, with configurable origin and direction
The ability to reorder between these schemes is crucial for compatibility with different HEALPix implementations and for certain operations.
This is the default RING order:
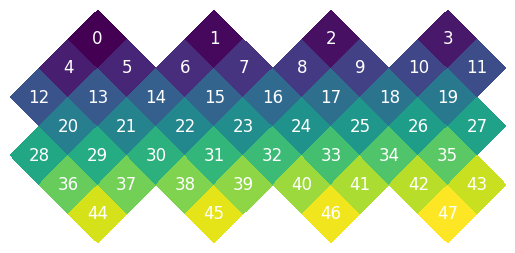
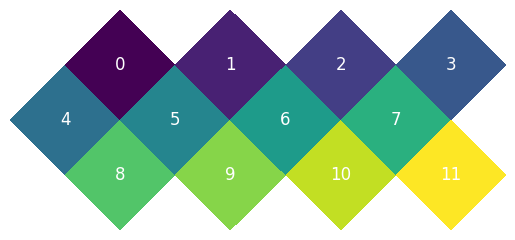
And the NEST
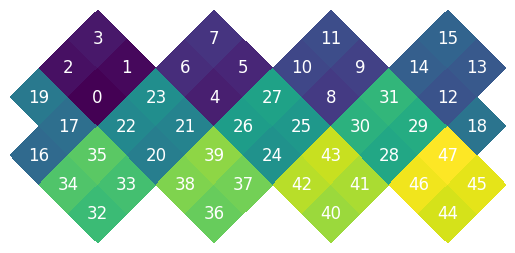
While NEST and RING are classic, we often want to treat each base pixel of the healpix grid as a 2d array. We have a XY pixel ordering object that allows us to do this.
(2.0, 6.0)
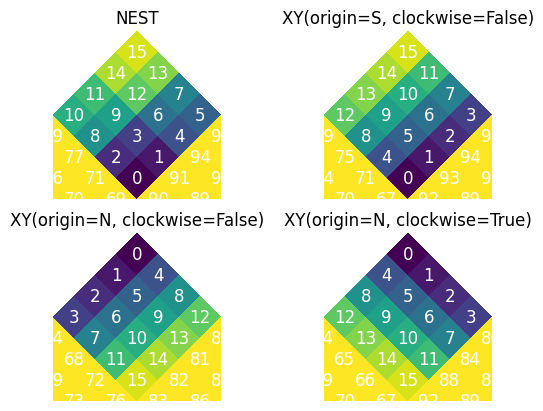
Why can convert between any two pixel orders like this:
healpix.reorder(x, healpix.PixelOrder.RING, healpix.PixelOrder.NEST) # ring to nest
healpix.reorder(x, healpix.PixelOrder.NEST, healpix.PixelOrder.XY()) # ring to XY
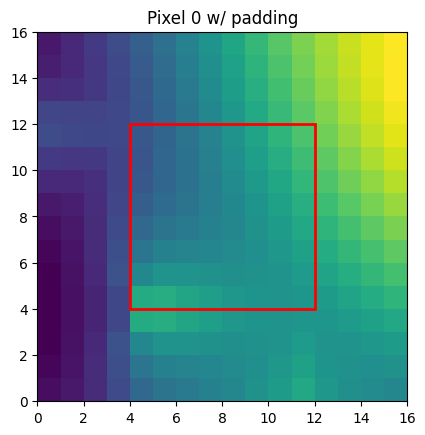
Padding and Convolutions#
The XY pixel order is useful for image analysis since it is a Cartesian-like grid. For example this is what the data looks like on each face of the sphere:
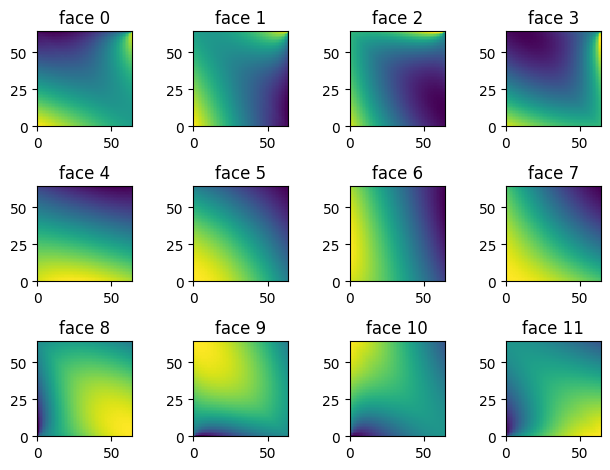
Padding is a basic primitive for many machine learning methods. For example, convolutions on the sphere can be implemented as a padding followed by a convolution. Because of this we have written a CUDA padding routine that works for the HEALPix grid. It requires (origin=N, orientation=clockwise) i.e. [N, E, S, W]. For convenience we have an alias for this healpix.HEALPIX_PAD_XY. Let’s now call the padding routine and plot the padded region.
nside = 8
pad_size = 4
# Let's assume some data on the NEST grid (like is common for datasets)
grid = healpix.Grid(level=healpix.nside2level(nside), pixel_order=healpix.PixelOrder.NEST)
field = np.cos(np.deg2rad(grid.lat+ 40)) ** 2 + 0.5 * np.sin(np.deg2rad(grid.lon))
field = torch.from_numpy(field)
# now reorder, reshape and pad
out = healpix.reorder(field, grid.pixel_order, healpix.HEALPIX_PAD_XY)
z = out.reshape([1, 12, nside, nside]) # healpix.pad requires a 4d input
z_padded = healpix.pad(z, pad_size)
/home/nbrenowitz/workspace/earth2grid/earth2grid/healpix/_padding/pure_python.py:131: UserWarning: Using a non-tuple sequence for multidimensional indexing is deprecated and will be changed in pytorch 2.9; use x[tuple(seq)] instead of x[seq]. In pytorch 2.9 this will be interpreted as tensor index, x[torch.tensor(seq)], which will result either in an error or a different result (Triggered internally at /pytorch/torch/csrc/autograd/python_variable_indexing.cpp:345.)
return x[slicers]
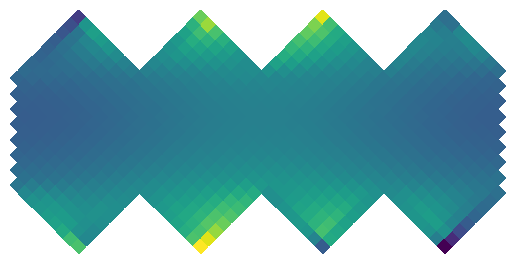
To run convolutions on the sphere
# convolution 3d is used to skip over the face dimension
conv = torch.nn.Conv3d(
in_channels=1, out_channels=1, kernel_size=[1, 3, 3], padding=0
)
pad_size = 1
b, c, f, n, n = 1, 1, 12, nside, nside
x = out.view(b, c, f, n, n).float() # some input data
# start processing
y = x
y = y.view(b * c, f, n, n) # combine (b, c) dims, healpix.pad requires a 4d input
y = healpix.pad(y, pad_size)
y = y.view(b, c, f, n+2 * pad_size, n+2 * pad_size)
y = conv(y)
print(x.shape, y.shape)
# reshape back for visualization
y = healpix.reorder(y.view(b, c, f * n * n), healpix.HEALPIX_PAD_XY, healpix.PixelOrder.RING)
healpix.pcolormesh(y[0,0].detach())
torch.Size([1, 1, 12, 8, 8]) torch.Size([1, 1, 12, 8, 8])
<matplotlib.collections.QuadMesh at 0x7c2ea4a83920>
Summary#
In this tutorial, we’ve covered several key aspects of working with HEALPix data:
Visualization: Using double pixelization to create visually appealing representations of HEALPix data while preserving the native pixel structure.
Zonal Averaging: Computing latitudinal averages to analyze patterns in the data.
Regridding: Transforming data between different grid types, including:
HEALPix to regular lat-lon grid
HEALPix to higher resolution HEALPix grid
Reordering: Converting between different HEALPix pixel ordering schemes:
RING: Pixels ordered in rings of constant latitude
NEST: Pixels ordered in a hierarchical, nested pattern
XY: Pixels ordered in a Cartesian-like grid within each face, with configurable origin and direction
Padding: Implementing efficient CUDA-based padding operations for machine learning applications, particularly useful for spherical convolutions.
These tools provide a comprehensive set of operations for working with global data on the sphere, making it easier to analyze, visualize, and process geophysical data for various applications including machine learning.