const char* a_query_string = "CGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTACGTA"
const uint32 query_string_len = strlen( a_query_string );
string_to_dna( a_query_string, query_string_len, h_query_string.begin() );
q, 2u,
n_query_qgrams,
d_query_indices.begin(),
d_query_qgrams.begin() );
thrust::sort_by_key(
d_query_qgrams.begin(),
d_query_qgrams.begin() + n_query_qgrams,
d_query_indices.begin() );
d_query_qgrams.begin(),
d_query_qgrams.begin() + n_query_qgrams,
d_ranges.begin(),

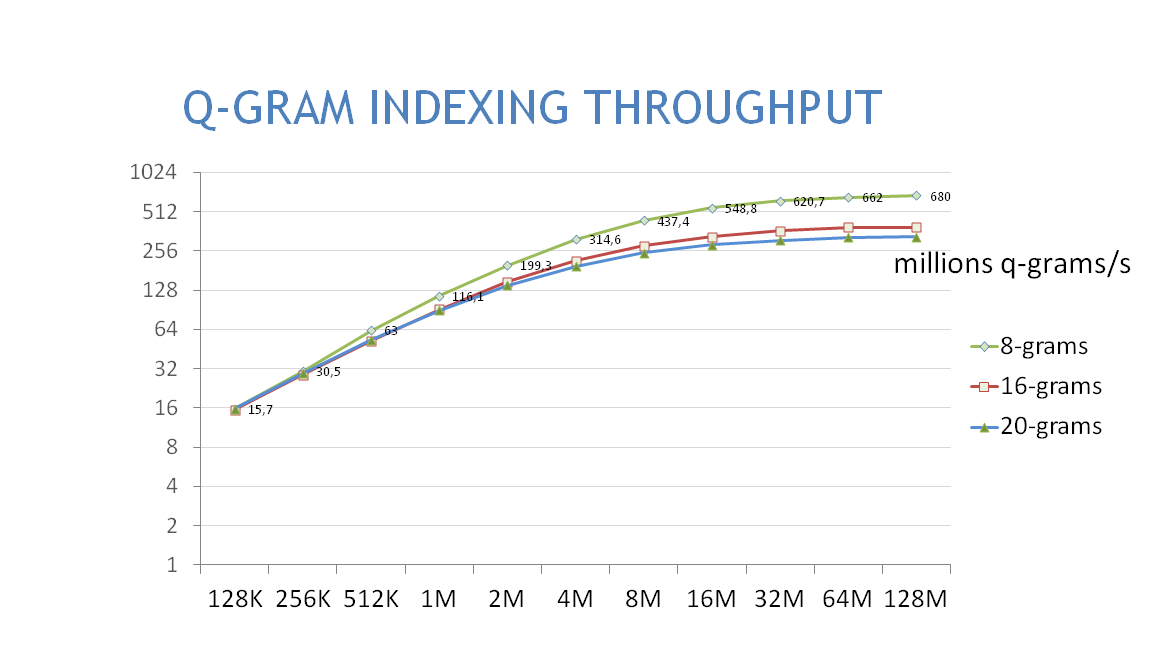
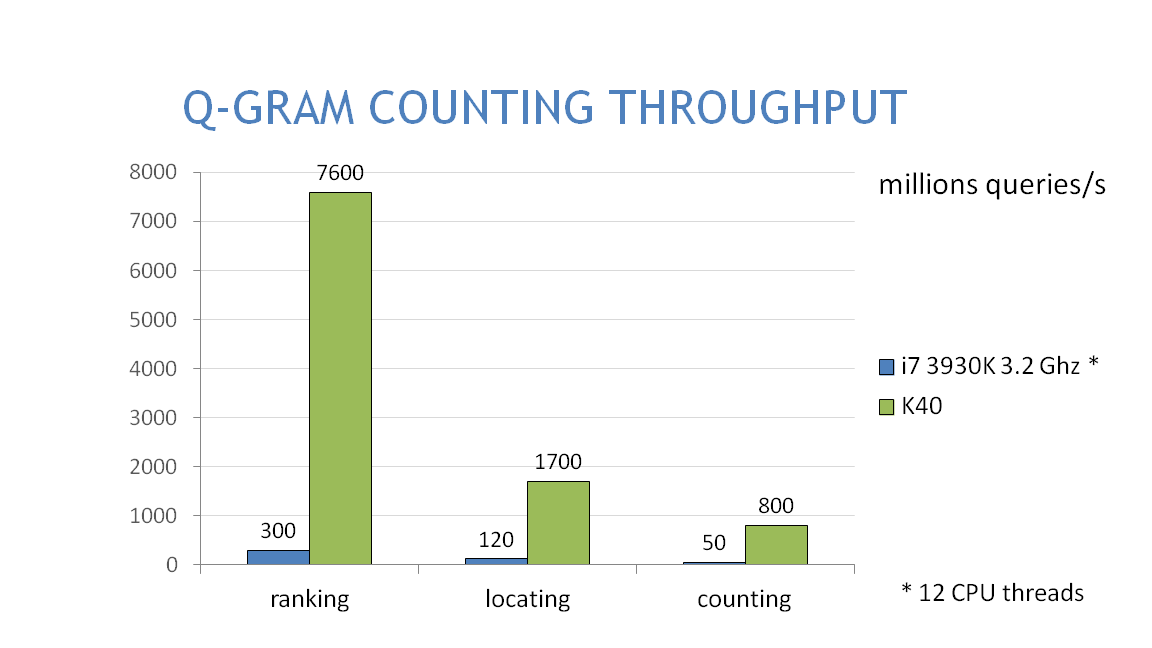
 1.8.4
1.8.4